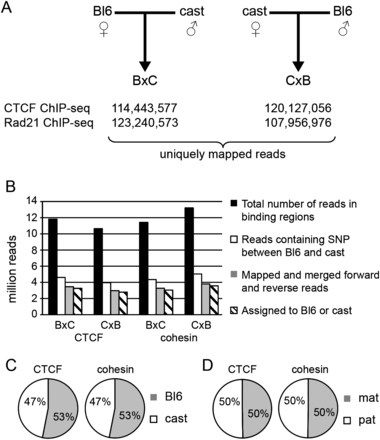
(A) ChIP-seq was performed for CTCF and cohesin (RAD21) on P21 brain in B × C and C × B F1 hybrid animals. The experimental design and number of uniquely mapped reads taken forward for further analysis are shown. (B) Regions of CTCF and RAD21 binding were identified using the Useq, and regions identified with a FDR of <13 were considered significant and were tested for parent-of-origin-specific binding. (Black bar) The number of reads for each experiment that fell at, or within ±500 bp of a binding region; (white bar) indicates reads in binding regions that aligned over a SNP between C57BL/6 (Bl6) and Mus m. castaneus (cast); (gray bar) the number of reads after the paired reads are considered together and the best-quality read is used to map the read to Bl6 or cast. (Hatched bar) The final number of reads assigned. There was a consistent bias toward the reference sequence (C); however, this effect was eliminated after we combined B × C and C × B reads (D).











