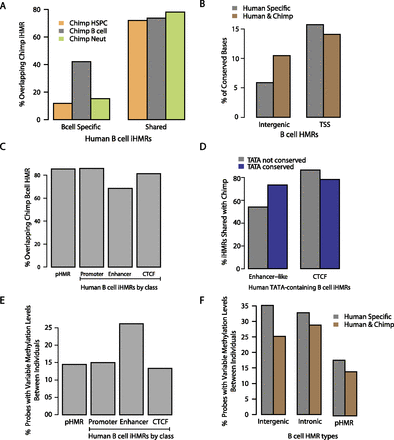
iHMRs are conserved in methylation state and sequence, and are enriched for human population variation in methylation levels. (A) Cell-type-specific hypomethylation is conserved between human and chimp. Overlap between human B cell–specific or shared HMRs with HMRs in different chimpanzee cell types. (B) Intergenic HMRs shared between human and chimp are also more conserved at the sequence level. (C) Enhancer-like iHMRs are more variable between human and chimp. Barplots show the percentage of B cell iHMRs of different classes that overlap a chimp B cell iHMR. (D) Conservation of the TATA motif at enhancer-like iHMRs predicts conservation of hypomethylation. Barplots show the percentage of human iHMRs (containing the TATA motif) shared with chimp for (left) transcribed, enhancer-like iHMRs and (right) CTCF iHMRs, depending on whether the TATA motif is conserved in chimp. (E) Methylation is more variable at enhancer-like iHMRs in the human population. Barplots show the fraction of probed loci in different HMR classes at which methylation levels vary significantly between whole-blood samples from individuals (see Methods). (F) iHMRs that are conserved with chimp are also less variable in methylation level between human individuals.











