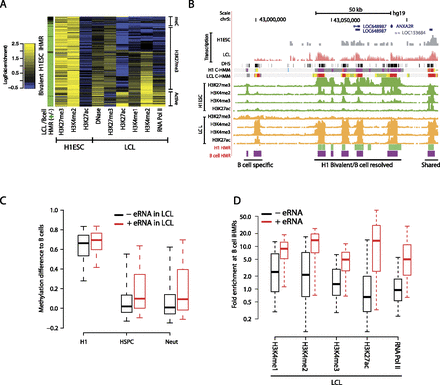
Resolution of bivalent iHMRs during differentiation and stepwise, de novo hypomethylation at transcribed enhancer-like iHMRs. (A) Bivalent iHMRs are resolved to active or silenced chromatin states during differentiation. Heatmap showing the LCL chromatin profile at iHMRs with a bivalent chromatin signature in H1 ESCs. Sidebar colors indicate whether the HMR remains hypomethylated in B cells (green) or becomes fully methylated (black). (B) Example locus surrounding ANXA2R, a gene expressed in lymphocytes and bone marrow, illustrating the coordinated resolution of the H1 ESC bivalent chromatin state in B cells/LCL. ENCODE regulation and transcription tracks are shown along with chromatin states modeled by ChromHMM (Ernst et al. 2011) in H1ES and GM12878 cells. Transcription tracks are presented in log scale. (C) Transcribed, active enhancer-like iHMRs in B cells are fully methylated in H1 ESCs and show intermediate states in other blood cell types. Differences in methylation levels between other cell types and B cell iHMRs are shown. (D) Only B cell iHMRs with eRNA (>0.1 RPKM) show strong enrichment for chromatin marks, suggesting an active regulatory state.











