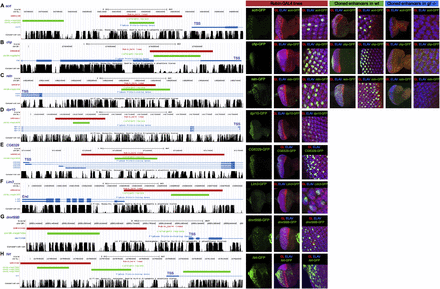
Validation of predicted Glass target enhancers by in vivo enhancer-reporter assays. Predicted enhancers with Glass binding sites were tested for nine genes, namely, scrt (A), chp (B), retn (C), dpr10 (D), CG6329 (E), Lim3 (F), dmrt99B (G), and Nrt (H). (Left) UCSC Genome Browser screenshots showing the location of the predicted Glass target CRM by cisTargetX (green) and the overlapping enhancer-GAL4 lines from Janelia Farm (red). For the top three enhancers, the green regions were cloned in front of GFP and tested by transgenesis (see Methods). For the remainder, only the red region is tested by crossing the Janelia Farm GAL4 line with UAS-GFP. (Right) Immunohistochemistry using anti-GFP (green) showing the activity of the tested region, together with anti-Glass (red), the predicted regulator (expressed in photoreceptors), and anti-Elav (blue), a general neuronal (here photoreceptor neuron) marker. (Blue column) The complete loss of activity of the cloned enhancer in a homozygous glass mutant background.











