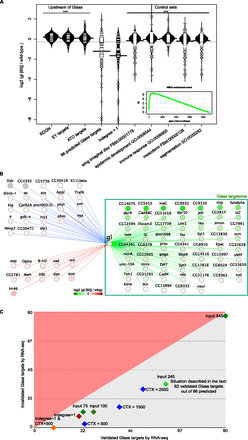
Validation of predicted Glass target genes by RNA-seq. (A) Bean plots representing the log2 (gl[60j]/wild-type) for the 96 predicted Glass target genes, compared with the same values for control gene sets. As control sets we used three sets of genes expressed upstream of glass, namely, all genes from the Retinal Determination Gene Network (RDGN) from Amore and Casares (2010); Eyeless target genes from Ostrin et al. (2006); and Atonal target genes from Aerts et al. (2010). We also used five other negative control sets, unrelated to eye development. (***) Significant difference (Wilcoxon FDR <0.05), compared with the Glass target sets. (Inset) Gene Set Enrichment Analysis (GSEA) showing a significant (FDR <0.001) enrichment of the 96 genes at the top of the ranking. (B) The predicted Glass targetome with 96 genes, showing overall down-regulation (green). (Dashed red edges) Up-regulation; (blue edges) no expression changes in the mutant. (C) Comparison of the amount of validated, or true-positive Glass target predictions (x-axis; genes that are down-regulated in the glass mutant) and invalidated, or false-positive predictions (y-axis; genes that are not down-regulated in the mutant). All situations show a higher number of true positives than false positives. (Blue diamonds) Different filters based on the cisTargetX genomic ranking; (green diamond) the situation in the text (62/96 validated targets); (orange diamond) a very stringent filtering with 100% positive predictive value, although retaining only eight Glass target genes.











