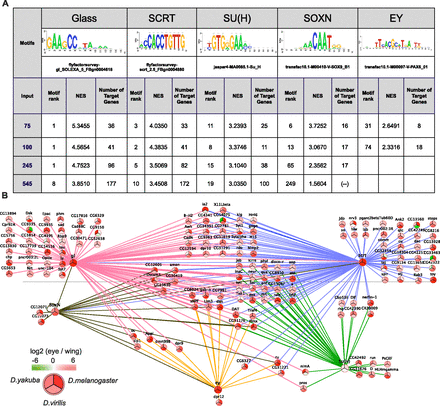
Motif discovery results from cisTargetX. (A) Motifs and candidate TFs found by cisTargetX, using different gene set sizes as input (top 75, 100, 245, and 545 genes from the OS-based integrative ranking). (NES) Normalized enrichment score from cisTargetX; (Motif Rank) rank of the motif from a collection of 3731 motifs used by cisTargetX. (B) Gene regulatory network showing the predicted regulatory interactions of Glass, SCRT, SU(H), SOXN, and EY, obtained from the cisTargetX analyses on the top100 and the top245 eye-specific genes. (Dashed arrows) Glass target gene predictions that are found to be up-regulated in the glass mutant by RNA-seq.











