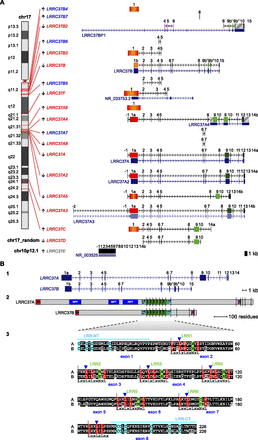
(A) LRRC37 family organization in human. The structure of complete and partial LRRC37 genes according to the reference human genome (hg18) is shown. (Left) Chromosomal band location of each locus on human chromosome 17 ideogram (only LRRC37E maps to chromosome 10 [gray] and is a retrocopy; LRRC37D does not have a chromosomal assignment). Genes are indicated as expressed (red) or not expressed (blue) based on the analysis of EST (expressed sequence tag) data. (Right) Variation in LRRC37 structures. Exons are represented as blocks connected by horizontal lines with arrowheads depicting introns. Exon 1 of A, A2, A3, and A4 copies (1a, in red) is longer than exon 1 of the B copy (1b, in orange); the B and B2 copies have exon 9 sequence split into two exons, 9b′ and 9b″. Exon 1 with deletions and/or insertions is red-orange. Exon 9 is dark green; exon 9 with deletions and/or insertions is light green. LRRC37A3 copy has one additional exon located at 5′ but lacks exon 12, and exon 13 is not transcribed. LRRC37A4 copy lacks exons 6 and 7 but carries a tandem duplication of exons 9 and 10. LRRC37A5 has a deletion of a single nucleotide at the end of the exon 1 sequence, causing a frameshift of the reading frame and the formation of a stop codon at the end of exon 1. LRRC37B2 is a fusion gene: exons 7, 8, and 9 match exons 9b′, 9b″, 10, and 15 of LRRC37B, whereas exons 4, 5, and 6 derive from a duplication of exons 4, 5, and 6 of SMAD-specific E3 ubiquitin protein ligase 2 (SMURF2) (NM_022739) (in pink), which maps at 17q24.1. Exapted introns are gray striped. RefSeq genes annotated in these loci are reported, following the same display conventions as in the UCSC Genome Browser. (B) Human LRRC37A and LRRC37B gene and protein structures. (Top panel) LRRC37A and LRRC37B cDNA predict an ORF with the methionine start codon (exon 1) and a stop codon (exon 14 and exon 15, respectively). LRRC37A encodes a predicted protein of 1700 amino acids with a molecular weight of 188 kDa. LRRC37B encodes a predicted protein of 947 amino acids with a molecular weight of 106 kDa. (Middle panel) Predicted structure of human LRRC37A and LRRC37B proteins according to SMART and LRRscan tools. Signal peptides (red), LRR motifs (green), LRR-NT and LRR-CT domains (turquoise), transmembrane helices (pink), and other repetitive motifs not associated with a known domain (blue) are shown. Exon boundaries are indicated with vertical dashed lines. (Bottom panel) An alignment of LRR regions of human LRRC37A and LRRC37B. The six LRR motifs and the final LRR are shown with the conserved leucine and asparagine residues highlighted in red and green, respectively. Boundaries between LRR motifs are marked with vertical dashed lines. LRR-NT and LRR-CT domains are shown with the conserved cysteine residues highlighted in turquoise. Exon boundaries are indicated with blue arrowheads.











