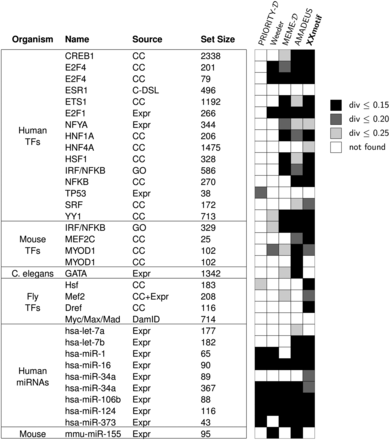
Top 4 benchmark results on 24 target sets for transcription factors from human, mouse, worm, and fly, as well as 10 target sets for microRNAs from human, and mouse from the metazoan target set compendium (Linhart et al. 2008). The plot is adapted from Linhart et al. (2008): The “Source” column indicates the experimental procedure or database from which the target set was derived: Gene-expression microarrays (Expr), ChIP-chip (CC), ChIP-DSL (C-DSL), DamID (van Steensel et al. 2001), or Gene Ontology (GO) database (Ashburner et al. 2000). The black and gray boxes indicate the similarity of the predicted PWM to the reference motif in TRANSFAC or miRBase. Darker shades indicate closer similarity. “Set Size”: number of sequences within the input set.











