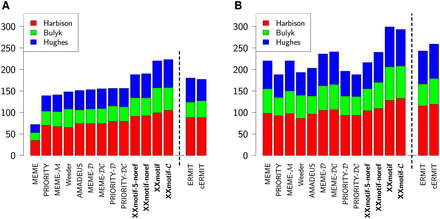
Sensitivity of motif discovery tools on yeast ChIP-chip data. Shown is the number of correctly predicted transcription factor binding motifs within the top 1 (A) or top 4 predictions (B). Predictions are based on ChIP-enriched intergenic regions from 352 ChIP-chip experiments (Harbison et al. 2004). Three experimental reference sets are used to judge the correctness of motifs (red, green, blue). The dashed line separates the general-purpose motif discovery tools from ERMIT, which needs ChIP enrichment P-values. In the tool names, M indicates a fifth-order Markov model, C the use of conservation, and D the discriminative prior from the Hartemink lab (Gordân et al. 2010). XXmotif-noref and XXmotif-5-noref omit the PWM refinement and the latter version uses only 5-mer seeds.











