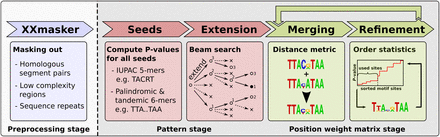
Overview of XXmotif with its three main stages. After an optional step to mask confounding sequence regions (blue), enrichment P-values of all 5-mers and gapped palindromic and tandemic 6-mer seed patterns are evaluated, and the best seeds are recursively extended by an optional gap and a motif position (red). Patterns are converted to PWMs and fed to the PWM stage (green). Here, similar PWMs are merged and then iteratively refined by optimizing the motif enrichment E-value. Finally, merging and refinement stages are iterated until convergence.











