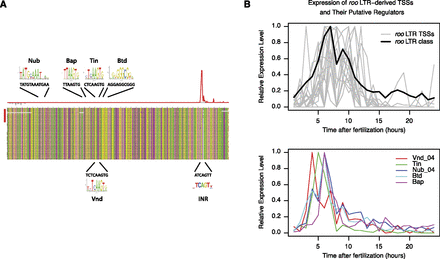
Core promoters and cis-regulatory elements in transposable elements: roo LTRs. (A) Multiple alignment of the sequences of the 18 LTRs providing TSCs for host genes (red bar on the left) to the roo consensus (upper sequence) and to a set of full-length LTRs with high similarity to the class consensus. The histogram above shows the density of tags on the upper (red) and lower (gray) strands. The positions of various sequence motifs are depicted, along with the logo of the known motif and the actual consensus sequence of the LTR. The TFBSs for NUB and BAP and the Initiator sequence (INR) are on the upper strand; the TFBSs for TIN, VND, and BTD are on the lower strand. (B) Expression profiles of the genes encoding putative regulators of roo LTRs. nub and vnd have more than one TSS, and only the one with the expression profile most consistent with roo LTRs is shown.











