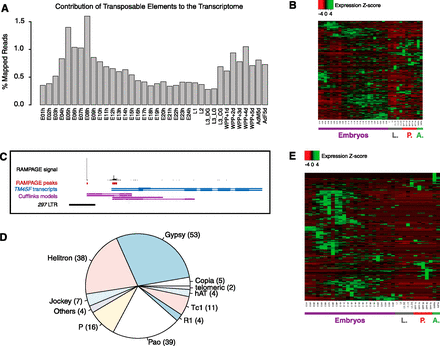
Transposable elements display developmental regulation and provide TSSs for many host genes. (A) Contribution of transcription initiating within transposable elements to the developmental transcriptome. For each time point, we report the proportion of all mapped reads (aligned uniquely or to multiple locations) for which the 5′ end lies in an annotated transposon. (B) Z-score–normalized expression profiles for all annotated classes of transposable elements. Note the developmental regulation of virtually all classes, as well as the disparity of patterns across classes. (C) A 297 LTR provides a strong alternative promoter for the TM4SF gene. (D) Subfamilies of transposable elements providing TSSs for annotated genes. The number of TSCs for each subfamily is reported in brackets (total 182). (E) Z-score–normalized expression profiles for all transposon-derived genic TSCs. The diversity of expression profiles underscores the versatility of transposons as regulatory modules.











