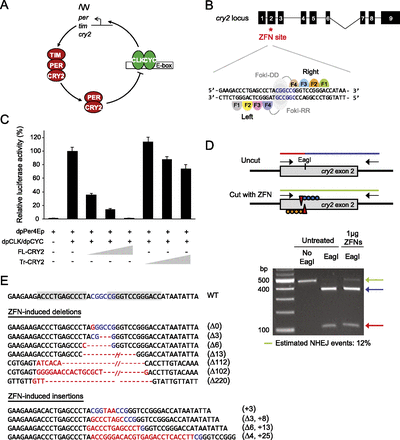
Validation of ZFN activity to target cry2 in DpN1 cells. (A) Proposed core transcriptional feedback loop of the monarch circadian clockwork. CLOCK (CLK) and CYCLE (CYC) heterodimers drive the transcription of period (per), timeless (tim), and cryptochrome 2 (cry2), which upon translation form complexes, cycle back into the nucleus, where CRY2 inhibits CLK:CYC-mediated transcription on a 24-h basis. (B, top) Schematic of monarch cry2 gene and position of the ZFN-targeted site (red star). (Black boxes) Exons. (Bottom) Magnified view illustrating binding sites for the ZFN pair, each consisting of four zinc-finger modules linked to either DD or RR variants of the FokI endonuclease. (C) Wild-type CRY2 (amino acids 1–742) represses CLK/CYC-mediated transcription in S2 cells in a dose-dependent manner, while the truncated protein (amino acids 1–160) does not. The monarch per E-box enhancer luciferase reporter (dpPer4Ep; 10 ng) was used in the presence (+) or absence (−) of dpCLK/dpCYC expression plasmids (5 ng each) and either the full-length CRY2 (FL-CRY2; 1, 2, and 10 ng) or truncated CRY2 (Tr-CRY2; 1, 2, and 10 ng). Luciferase activity is relative to beta-galactosidase activity and normalized so that the relative activation by dpCLK/dpCYC alone is 100%. Each value is the mean ± SEM of three independent transfections. (D, top) Strategy used to detect ZFN-induced mutations in monarch cells by restriction endonuclease assay. (Red and blue lines) Wild-type genomic amplicon showing the presence of an EagI site. Genomic fragments with mutations induced by NHEJ are resistant to restriction enzyme digestion because of the loss of the EagI site and appear uncleaved (green line). (Bottom) Estimation of ZFN activity in DpN1 cells. Genomic amplicons from a pool of cells untreated or treated with 1 μg of ZFNs, subjected to restriction digest. The frequency of NHEJ in treated cells was estimated by quantification of ethidium bromide staining and densitometry of the resistant band (green arrow) relative to wild-type fragments (blue and red arrows). (E) ZFN-induced cry2 mutations in DpN1 cells. (Gray shaded boxes) The ZFN recognition sites on the wild-type sequence. (Blue letters) The EagI site. The positions of (red dashes) deletions and (red letters) insertions.











