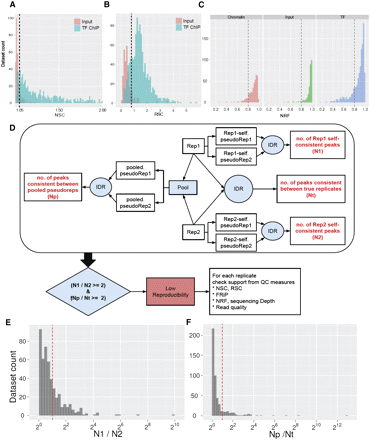
Analysis of ENCODE data sets using the quality control guidelines. (A–C) Thresholds and distribution of quality control metric values in human ENCODE transcription-factor ChIP-seq data sets. (A) NSC, (B) RSC, (C) NRF. (D) IDR pipeline for assessing ChIP-seq quality using replicate data sets. (E,F) Thresholds and distribution of IDR pipeline quality control metrics in human ENCODE transcription factor ChIP-seq data sets. (Dashed lines) Current ENCODE thresholds for the given metric, which are NSC > 1.05 (A); RSC > 0.8 (B); NRF > 0.8, N1/N2 ≥ 2 (where N1 refers to the replicate with higher N) (E); Np/Nt ≥ 2 (F).











