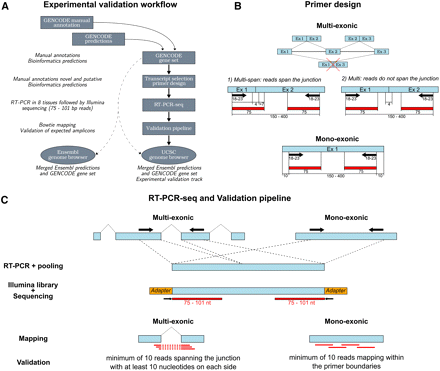
RT-PCR-seq workflow. (A) Schematic workflow of the experimental validation of GENCODE transcript models with RT-PCR-seq. (B) Position of primers (black arrow) designed to validate gene models and corresponding sequencing reads (red rectangles) on targeted exons (blue rectangles); (Ex) exon. We experimentally assessed three categories of gene models: (1) spliced models in which one primer could be placed within 75 nt of a junction and that will result in about half of the sequencing reads covering the junction (“Multi-span”); (2) spliced models in which this was unfeasible (“Multi”); and (3) monoexonic genes (“Mono”). Numbers below exons, primers, sequencing reads, and amplimers indicate their respective sizes. Exon–exon junctions skipping an annotated cassette exon were not considered (see example barred with a red cross). (C) Schematic representation of the criteria used to experimentally validate GENCODE gene models (see Methods for details and panel B for color code).











