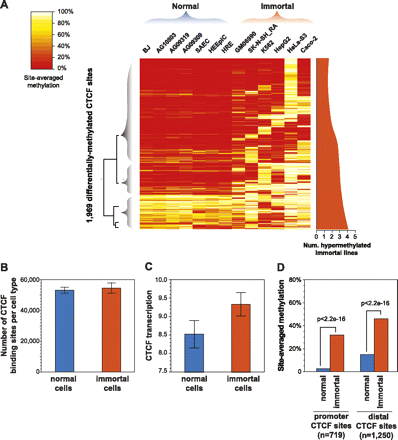
Cell-selective patterns of methylation associated with occupancy differences. (A) Methylation status at 1969 CTCF sites where differential methylation is significantly associated with occupancy differences. Color corresponds to the percentage of bisulfite sequencing tags at each site overlapping methylated CpG positions. Dendrogram (left) highlights pattern of hypermethylation in immortal cell lines. (Right) Smoothed plot of number of immortal lines exhibiting hypermethylation at each site. (B) Immortal lines show no significant difference in number of occupied CTCF sites (y-axis, mean). Error bars, SD. (C) immortal lines demonstrate increased CTCF transcript levels (y-axis, mean). Error bars, SD. (D) Immortal lines exhibit increased methylation relative to the other cell types, though significant promoter methylation is rarely observed in normal lines. y-axis, genome-wide median of per-site methylation. P-values, Wilcoxon. Promoter, ±2.5 kb of RefSeq transcription start site.











