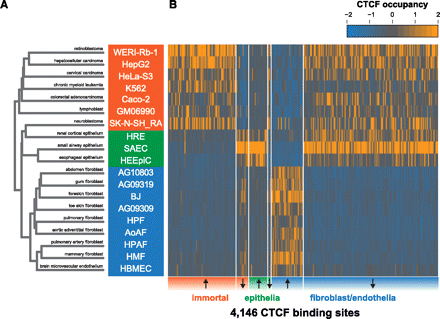
CTCF occupancy distinguishes similar cell types. (A) Unsupervised hierarchical clustering of binding at all CTCF sites. (B) CTCF occupancy at 4146 variable binding sites that distinguish immortal cell lines, epithelia, fibroblasts and endothelia (Methods). x-axis, CTCF binding sites in chromosomal order, separated into sites that are up-regulated and down-regulated (arrows) in each of the three groups (immortal, epithelial, fibroblast, and endothelial). Color corresponds to Z-score of normalized ChIP-seq density.











