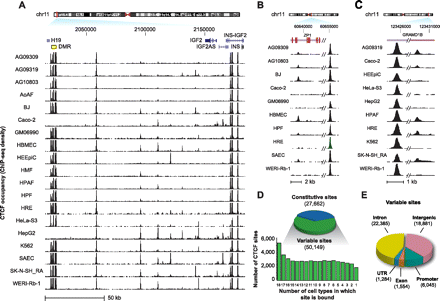
CTCF in vivo binding exhibits widespread plasticity. (A–C) Constitutive and variable CTCF sites. (A) The H19/IGF2 imprinted locus in multiple human cell types. Note the total silencing in two cell lines of the seven CTCF sites in the differentially methylated region (DMR; yellow box at left), and the complex pattern of cell-selective CTCF binding flanked by constitutive sites. Location (hg19), chr11:2,015,000–2,184,000. (B,C) Additional examples of variable sites. (D) Genome-wide analysis of CTCF binding in 19 cell types reveals 77,811 distinct binding sites; 27,662 sites are constitutively present in all cell types; 50,149 variable sites exhibiting a wide range of selectivity are present in a subset of one to 18 cell types (below). (E) Genomic distribution of variable sites is similar to constitutive sites (Supplemental Fig. S2A).











