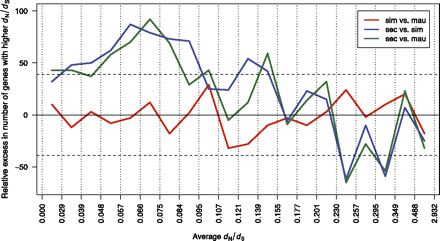
Reduced efficacy of selection in D. sechellia. A total of 7008 autosomal genes are binned according to level of selective constraint as measured by the average dN/dS between the inferred ancestor and each species. In each bin, three sign tests are performed to determine whether, for a given species pair, one species has significantly more genes with higher dN/dS. (Y-axis) Relative excess of genes with higher dN/dS for the first species in the pair. For example, in the first bin, D. simulans has 185 genes with higher dN/dS than D. mauritiana, while D. mauritiana has 175 genes with a higher dN/dS value. The difference, 10, is plotted. Values outside the region delimited by the dashed lines are significant for the sign test. D. sechellia tends to have higher dN/dS values in genes with high constraint (dN/dS < 0.155) and lower dN/dS in genes that experience positive selection or less constraint (dN/dS > 0.230). There is no significant difference between D. simulans and D. mauritiana for any of the bins.











