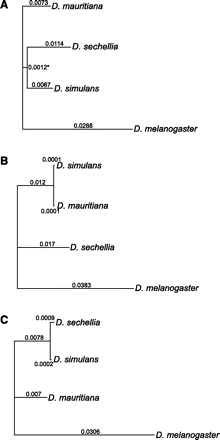
Sampling of phylogenetic trees. Three maximum likelihood trees with branch lengths that are estimated under a GTR model of nucleotide substitution. The trees are reconstructed from all aligned sites in the genome (A), all aligned sites from the mitochondrial genome (B), and a 15-kb region on chromosome 3R (C) (D. melanogaster reference coordinates 3R: 16,675,000–16,690,000) that includes the Ir93a and CG3822 ionotropic glutamate receptor genes. The asterisk on the interior branch in panel A indicates that this branch length can be regarded as statistically greater than zero; the resulting node has 100% bootstrap support.











