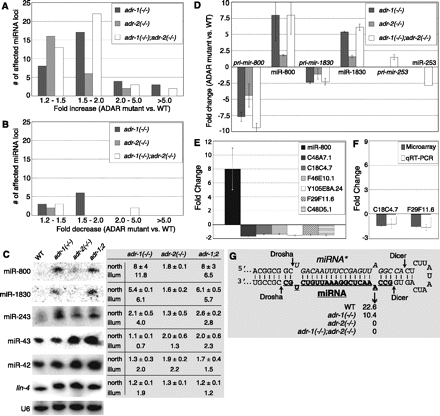
ADARs affect miRNA and mRNA target levels, but few miRNAs are edited. (A,B) Number of loci with increased (A) or decreased (B) reads in ADAR mutants compared to WT were plotted on the y-axis. miRNA loci were binned (x-axis) according to their fold-change in a mutant strain compared to WT. All miRNA loci altered ≥2-fold were affected in at least one mutant strain; loci altered ≥1.2-fold but <2-fold were affected in at least two mutant strains. (C) Representative Northern blot of mirVana enriched RNA (2 μg) from indicated strains, probed for miRNAs, with U6 as a loading control. Fold-change determined by sequencing (illum) and Northern blot (north) were tabulated, comparing each mutant to WT. Blanks indicate sequencing did not predict a significant difference. (Error) Standard deviation (STD), adr-1(−/−);adr-2(−/−) abbreviated adr-1;2. (D) Fold-change of pri-miRNA in ADAR mutants compared to WT, as determined by qRT-PCR, was compared to change in miRNA. Levels of miR-800 and miR-1830 were determined by Northern blot, miR-253 by sequencing; the latter did not show a significant change in single mutants. (E) Fold-change of miR-800 (see Fig. 1C, Northern blot) compared to fold-change of predicted mRNA targets, as determined by microarray analyses (adr-1[−/−];adr-2[−/−] vs. WT worms) (Supplemental Table S2). (Error bars) Standard error of the mean (SEM) for mRNAs, STD for miR-800. (F) qRT-PCR validation of miR-800 mRNA target levels in adr-1(−/−);adr-2(−/−) worms compared to WT. (Error bars) SEM for microarray, STD for qRT-PCR. (G) Structure of pri-mir-800, with miRNA sequence underlined and bold and miRNA* sequence italicized. Percent editing at indicated adenosine (double arrowhead) is tabulated for each strain. (Single arrowheads) Drosha and Dicer cleavage sites.











