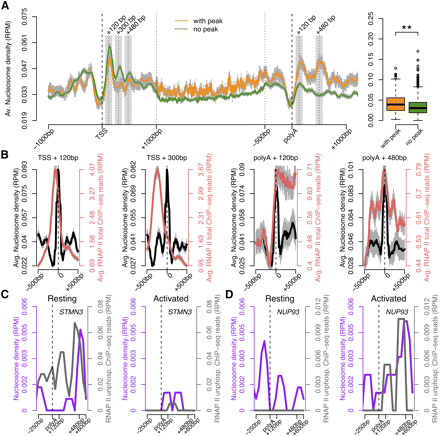
Nucleosome organization correlates with RNAPII density at the 3′-end. (A) Average nucleosome occupancy across genes with (orange) and without (green) a peak of total RNAPII at the 3′-end. The y-axis shows the normalized number of sequence tags of DNA at each position. The 5′- and 3′-ends are unscaled. The remainder of the gene is rescaled to 2 kb. The boxplot represents the average nucleosome occupancy for each gene in the termination region [poly(A) site to 500 bp downstream]. (**) P-value <0.00005 by two-sided Mann-Whitney test. (B) Average nucleosome distribution (black) and total RNAPII density (red) around the position of maximum nucleosome occupancy (0) at the indicated regions. Error bars (gray), SEM. (C,D) Nucleosome (purple) and RNAPII (gray) occupancy along two genes, the expression of which is either down- (STMN3, C) or up-regulated (NUP93, D) in human CD4+ T cells activated by TCR signaling. (RPM) Reads per million.











