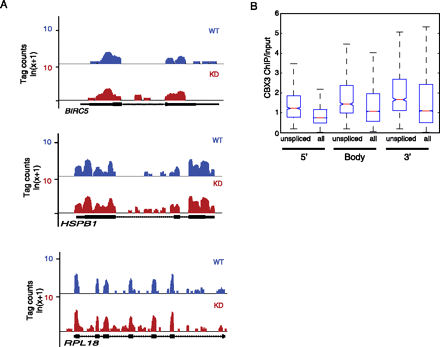
Loss of CBX3 causes RNA processing defects genome-wide. (A) UCSC Genome Browser snapshots showing representative RNA-seq data of CBX3 target genes BIRC5, HSPB1, and RPL18. Expression tag counts in ln(x + 1) scale. Gene schematic showing exons (filled black box) and introns (hashed lines) below tracks. (B) ChIP-seq CBX3 enrichment (IP/input RPKM) at 5′end, coding region, and 3′ end of genes having increased intron inclusion (unspliced) in KD versus WT HCT116 cells as compared to average CBX3 enrichment for all genes. Expression of introns and exons analyzed by RNA-seq in WT and KD cells. For box plots, the median is represented by the red line, the edges of the box represent the 25th and 75th percentiles, and the whiskers extend to non-outlier extreme data points.











