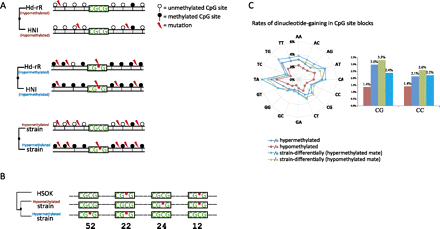
(A) Representative mutation patterns in evolutionarily conserved hypo- and hypermethylated regions and in regions that are differentially methylated in different strains, where substitution rates ascend from top to bottom. The CGCG motif is significantly conserved in hypomethylated regions (P < 10−680 by a two-proportion z-test). (B) Number of strain-differentially methylated CpG site blocks with either gain or loss of the CGCG motif. Of 1656 CGCG motif occurrences in multiple alignments of the genomes of the three strains, 52 (24, respectively) were conserved in hypomethylated (hypermethylated) regions of one of Hd-rR or HNI but were mutated in hypermethylated (hypomethylated) regions of the other strain. We then evaluated the significance of the difference between the means in the two groups, 52/1656 vs. 24/1656, to obtain a P-value of 0.203% according to a two-proportion z-test. (C) The rates of dinucleotide gain in CpG site blocks. Gain rates in the hypo- or hypermethylated CpG site blocks and each mate in strain-differentially methylated blocks are shown.











