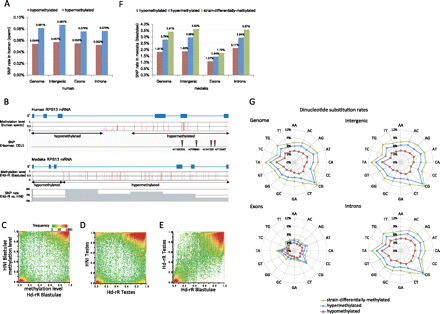
Methylation patterns and substitution rates in the inbred medaka strains, Hd-rR and HNI. (A) SNP (single-nucleotide polymorphism) rates in hyper- and hypomethylated CpG blocks in the reference human genome (hg19). The difference in SNP rates was significant in the entire genome (P < 10−566 by a two-proportion z-test) (Supplemental Table S4), in intergenic regions (P < 10−305), in exons (P < 10−29), and in introns (P < 10−151). (B) Methylation level and SNP distribution in the homologous regions of the human and medaka genomes where gene RPS13 is coded. (C,D) Comparisons of the methylation patterns in Hd-rR and HNI. The vertical and horizontal axes indicate methylation level. The heat map uses logarithmic coordinates and presents the number of corresponding CpG site blocks. Conserved hypermethylated and hypomethylated patterns between the two strains were dominant, except for a small number of hot spots observed in the differentially methylated regions (differences in methylation level ≥ 0.5). (E) Comparison of the methylation patterns in blastulae and testes in Hd-rR. (F) SNP rates in hypo-, hyper-, and strain-differentially methylated regions in medaka blastulae grouped by the entire genome, intergenic regions, exons, and introns. The differences between SNP rates of hypo- and hypermethylated regions were remarkable: P < 10−2170 (genome), P < 10−2170 (intergenic regions), P < 10−113 (exons), and P < 10−589 (introns) according to a two-proportion z-test (Supplemental Table S4). Furthermore, the differences between SNP rates of strain-differentially and hypermethylated regions were also significant (Supplemental Table S4). (G) Dinucleotide substitution rates in the whole medaka genome, intergenic regions, exons, and introns in CpG site blocks with various methylation states. Color key presents mutation rates: blue for hypermethylated (methylation level ≥ 0.8 in both strains); red for hypomethylated (methylation level ≤ 0.2 in both strains); and green for strain-differentially methylated (difference in methylation level between the two strains ≥ 0.5) in blastulae. The axes in each radar chart represent substitution rates of individual dinucleotides. Each dinucleotide shows the same substitution rate as its reverse complementary dinucleotide. Significant differences between substitution rates in hypo- and hypermethylated regions were observed for all dinucleotides, and the P-values, according to a two-proportion z-test, were P < 10−441 (genome), P < 10−263 (intergenic regions), P < 10−15 (exons), and P < 10−69 (introns) (Supplemental Table S5).











