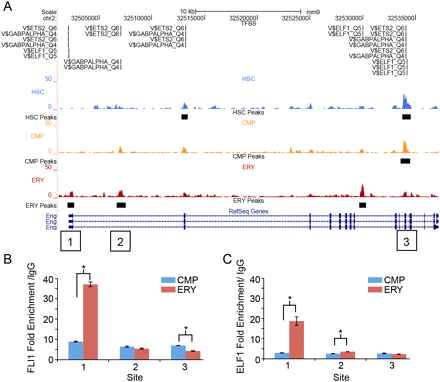
FLI1 and ELF1 occupancy of methylation peaks in the Eng gene. (A) UCSC Genome Browser view of the Eng locus on chromosome 2. Transcription factor consensus sites (TRANSFAC IDs) are indicated above the MBD-seq data in regions of significant peaks. QPCR data verifying enriched ChIP binding of FLI1 (B) and ELF1 (C) at three sites of methylation peaks throughout Eng. Site 1, an ERY-specific peak, is within the proximal promoter; site 2, an ERY-specific intronic peak; and site 3, a progenitor (HSC and CMP) peak in the 3′ coding region. Comparison of binding in CMPs and ERYs revealed statistically significant differences of P = 0.004 (site 1, FLI1), P = 0.009 (site 1, ELF1), P = 0.021 (site 2, ELF1), and P = 0.012 (site 3, FLI1).











