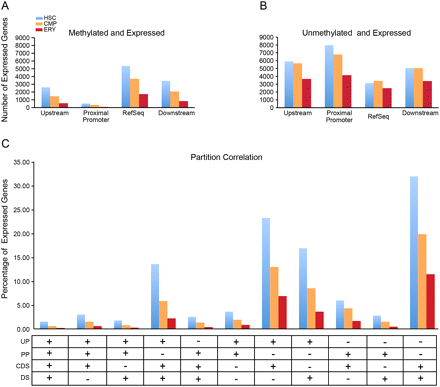
Methylation in genic partitions of expressed genes. Gene expression was obtained from the BloodExpress (Miranda-Saavedra et al. 2009) gene expression database for each cell type and compared with MBD-seq peaks. (A) The number of expressed genes (y-axis) in each cell type with methylation peaks in the distal promoter, proximal promoter, RefSeq gene body, and downstream region. (B) The number of expressed genes (y-axis) lacking methylation peaks in each of the genic partitions for each cell type. (C) Combinatorial analysis of methylation peaks occurring in all four genic partitions in expressed genes. (UP) Upstream 10 kb; (PP) proximal promoter; (CDS) RefSeq gene boundary; (DS) downstream. The presence (+) or absence (−) of methylation in the indicated partition is shown below the x-axis. The y-axis shows the percentage of expressed genes with each methylation pattern.











