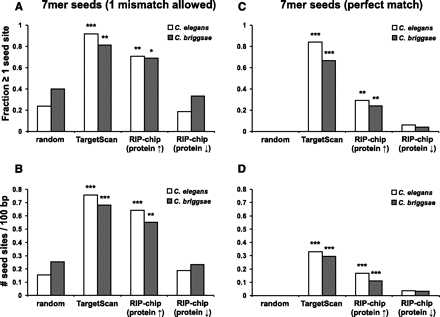
Targets identified by RIP-chip-SRM are enriched in evolutionarily conserved bantam/miR-58 seed binding sites. The same data sets as in Figure 3A (bantam/miR-58 targets identified by RIP-chip, targets predicted by TargetScan, and random control group) were tested for the presence (A,C) and frequency (B,D) of imperfect (one mismatch allowed, A,B) and perfect (C,D) bantam/miR-58 7-mer seed binding sites in their 3′ UTRs and the predicted 3′ UTRs of their corresponding C. briggsae homologs. The RIP-chip candidates are shown subdivided into the “RIP-chip (Protein ↑)” (log2 proteinmir-58/wt > 0) and “RIP-chip (Protein ↓)” (log2 proteinmir-58/wt < 0) groups (as in Fig. 3C). P-values for the presence of bantam/miR-58 seed binding sites (A,C) were determined by a Fisher’s exact test (one-sided, with the random group as the null hypothesis). P-values for the frequency of bantam/miR-58 seed binding sites (B,D) were determined by hypergeomtric distribution (using the frequency of binding sites in the whole 3′ UTR-ome as the null hypothesis). (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.











