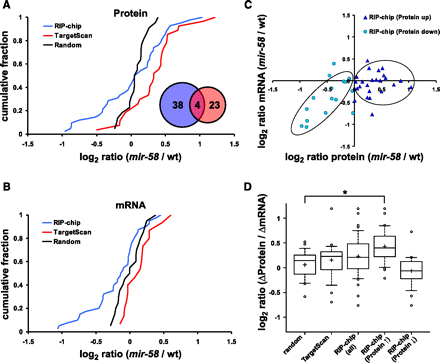
Bantam/miR-58 targets identified by RIP-chip are regulated mainly at the translational level. (A,B) Cumulative fraction plot of the log2 protein (A) and mRNA (B) changes (“mir-58” to “wild type”) of potential bantam/miR-58 targets identified by RIP-chip or TargetScan prediction and of a random control group. (A, inset) Overlap in membership between the RIP-chip and TargetScan groups. (C) Log-log plot of the protein (x-axis) and mRNA (y-axis) changes shown in A and B for the RIP-chip group. The RIP-chip candidates were subdivided into “RIP-chip (Protein up)” (log2 proteinmir-58/wt > 0; dark blue triangles) and “RIP-chip (Protein down)” (log2 proteinmir-58/wt < 0; light blue circles) groups, based on whether their protein levels were elevated or reduced in “mir-58” mutants. The circled data points represent the general correlation trend of the majority of data points within both groups. (D) Comparison of the extent of change in abundance at the protein and mRNA levels (proteinmir-58/wt/mRNAmir-58/wt) are depicted as box plots. The box plot marked with “*” differs significantly (P-value = 0.009, Kolmogorov-Smirnov test) from the random gene group. The RIP-chip candidates are also shown subdivided into the “RIP-chip (Protein ↑)” (log2 proteinmir-58/wt > 0) and “RIP-chip (Protein ↓)” (log2 proteinmir-58/wt < 0) groups. The bottom and the top of the boxes are the 25th and 75th percentile, respectively, while the whiskers represent the ninth and 91st percentile. The band within the box depicts the 50th percentile (median) and the “+” the mean value.











