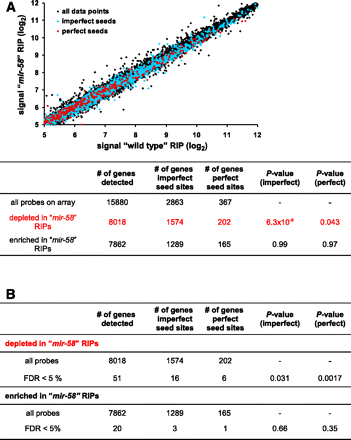
Identification of bantam/miR-58 targets by differential RIP-chip. (A) TAP::ALG-1 associated mRNAs were purified from synchronized L4 stage transgenic “wild-type” animals (WS4303; see Methods) and synchronized L4 stage transgenic “mir-58” mutants (WS5041; see Methods) and analyzed by one-color microarrays (Affymetrix). The average log2 expression values of all the detected mRNAs are shown. The abundance of TAP::ALG-1-associated mRNAs correlated strongly between both samples (Pearson square correlation factor [R2] = 0.99). Potential bantam/miR-58 targets are indicated in red (perfect 7-mer binding site in the 3′ UTR) and light blue (7-mer, one mismatch allowed). These potential bantam/miR-58 target genes were significantly overrepresented among the genes depleted in the “mir-58” RIPs, as expected for potential bantam/miR-58 targets. (B) Genes containing potential bantam/miR-58 binding sites were overrepresented among genes that were significantly depleted in the “mir-58” RIPs (FDR < 5%, SAM), as expected for potential bantam/miR-58 target genes. In contrast, no such overrepresentation could be detected for genes that were significantly enriched in the “mir-58” RIPs (FDR < 5%, SAM). P-values were determined by hypergeometric distribution.











