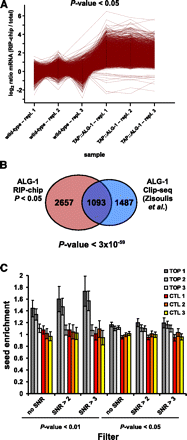
TAP::ALG-1 RIP enriches for miRNA targets. (A) RIP-chip of TAP::ALG-1-associated mRNAs. The log2 ratio profiles of the 4071 oligo probes (representing 3750 gene models, out of a total of 15,322 oligo probes, representing 12,890 gene models, detected) that were significantly changed (P-value < 0.05, two-sample t-test) in the TAP::ALG-1 RIP compared to the mock RIP are shown. Log2 ratios (RIP mRNA/total mRNA) were calculated for each measured oligo probe and normalized to the corresponding mock average log2 ratio for better visualization. A total of 99.7% of all features are enriched in the TAP::ALG-1 RIP. Three independent biological replicates were analyzed. Genotypes used: TAP::ALG1: alg-1(tm492); opIs205, wild type: N2 (= mock control). (B) The set of 3750 TAP::ALG-1-associated mRNAs identified here overlaps strongly (P-value < 3 × 10−59; hypergeometric distribution) with a previously published set of ALG-1-associated mRNAs identified by Clip-seq (Zisoulis et al. 2010). Only Clip-seq mRNAs that had also been detected on our microarrays (2560 out of 3093) were included in the analysis. The total number of expressed genes (12,890) on our microarray was used for the statistical analysis. (C) TAP::ALG-1 RIP enriches for mRNAs with seed binding sites for abundant miRNAs. miRNA seeds were arranged in groups of five based on their miRNA microarray expression values (see Supplemental Table S4). TOP1 includes the five most highly expressed miRNA seeds (including bantam/miR-58), TOP2 includes the next five most highly expressed seeds, etc. The CTL1, CTL2, and CTL3 groups each contained five miRNA seeds that were below the detection limit on the miRNA microarray (all seed groups are listed in Supplemental Table S13). Relative seed binding site enrichment in the 3′ UTRs of TAP::ALG-1-associated mRNAs compared to nonassociated mRNAs was determined for each group as described in Methods. Two different P-values and three different signal-to-noise ratio (SNR) cutoffs were applied to define the TAP::ALG-1 associated mRNAs.











