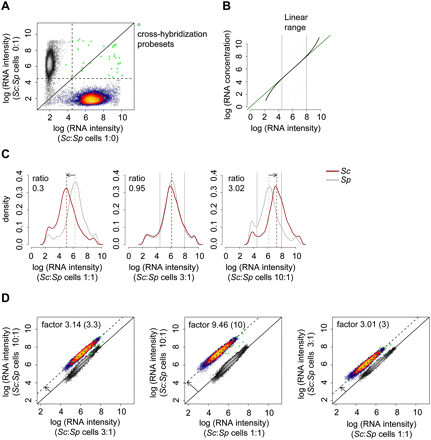
Establishing the cDTA protocol. (A) Assessment of cross-hybridization. Scatterplot of log intensities of 10,928 Affymetrix probe sets. The values on the x- resp. y-axis are obtained as the mean of two pure Sc resp. Sp replicate samples that were hybridized to the arrays. Sc and Sp probe sets (heat colored and gray scaled, respectively) can be separated almost perfectly. A total of 23 out of 5771 Sc probe sets show intensities above a (log) background intensity threshold of 4.5 in the Sp sample, whereas eight out of 5028 Sp probe sets were above background in the Sc sample. These 31 probe sets are regarded as affected by cross-hybridization (green circles). Of these, 16 probe sets were excluded from analysis because all probes were affected by cross-hybridization (Methods). (B) Linear measurement range. Exemplary illustration showing that the relation of mRNA concentration (real amount) and mRNA intensity (fluorescent scanner readout) follows the Langmuir adsorption model (Hekstra et al. 2003; Held et al. 2003, 2006; Skvortsov et al. 2007). The green line indicates linearity. (Black line) Sigmoidal behavior, resulting from noise at low-hybridization levels and saturation effects at high-hybridization levels. (Gray stripe) Linear measurement range that we defined as an intensity range of 4.5–8 (natural log basis) based on noise signals below 4.5, for example, for probes that detect transcripts of genes that were knocked out and based on observed saturation effects above 8. (C) Calibration of Sc:Sp cell mixture ratio. The optimal cell mixture ratio has been chosen to maximize the number of probes for both Sc and Sp that fall into the linear measurement range (B). Sc and Sp cells were mixed in Sc:Sp ratios of 1:1, 3:1, and 10:1. The respective median mRNA level ratios are 0.3, 0.95, and 3.02. Log (RNA intensity) distributions of Sc (red) and Sp (gray) are shown. The median intensity level of Sp is approximately three times higher than that of Sc. As a consequence, a Sc:Sp cell mixture ratio of 3:1 was used. (D) Comparison of the three different cell mixtures of (C) in pairwise log–log scatter plots. All arrays are normalized to a common median of 4052 Sp probe sets (gray-scaled). A total of 4475 Sc probe sets (those in the linear measurement range) are shown in heat colors. The parallel offset of the Sc probe sets from the main diagonal measures the mRNA level differences of Sc in the three cell mixtures. The differences should be 3.3, 10, and 3 when we plot Sc:Sp ratios of 10:1 vs. 3:1, 10:1 vs. 1:1, and 3:1 vs. 1:1, respectively. The corresponding measured offsets are 3.14, 9.46, and 3.01, and thus in very good agreement.











