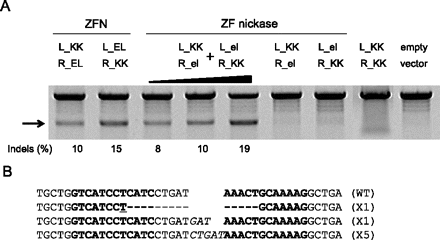
Two pairs of ZF nickases produce a DSB in the genome. (A) Nuclease or nickase-driven indels detected by T7E1 assay. PCR products amplified using genomic DNA from cells transfected with plasmids (4 μg/monomer) encoding nickases or nucleases were subjected to T7E1 digestion and analyzed by agarose gel electrophoresis. (Arrow) The expected position of the resulting DNA band. (The black triangle above the gel picture) The increase of the transfected plasmid (4, 8, and 10 μg/each monomer). (B) DNA sequences of the CCR5 wild-type and mutant clones. The two half-sites are shown in boldface letters. Microhomologies are underlined, and inserted bases are shown in italics. Dashes indicate deleted bases. The number of occurrences is shown in parentheses; X1 and X5 are the number of each clone. (WT) Wild type.











