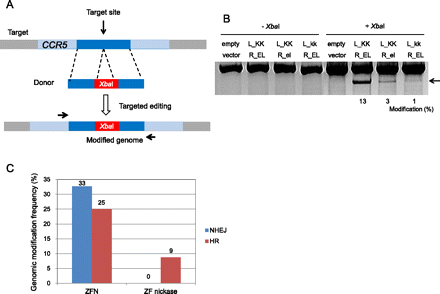
Targeted genome editing via HR in human cells with ZF nickases. (A) Schematic overview of HR-mediated genome editing. HR donor DNA consists of two 800-bp homology arms (left and right) and an XbaI site. K562 cells were transfected with plasmids encoding nucleases or ZF nickases plus HR donor plasmid. After 4 d of incubation, genomic DNA was isolated, and the target locus was amplified with primers (arrows) that bind outside of the homology arm sequences. PCR amplicons were digested with XbaI. (B) XbaI-treated and untreated DNA samples were analyzed on a 1% agarose gel. (Arrow) The expected position of XbaI-digested PCR products. Modification frequencies (percentages) were calculated by measuring the band intensity. (C) Comparison of patterns of genomic modifications induced by ZFNs and ZF nickases. PCR products corresponding to genomic modifications were cloned, sequenced, and classified according to their mutation patterns. The DNA sequence of each clone is shown in Supplemental Figure 1.











