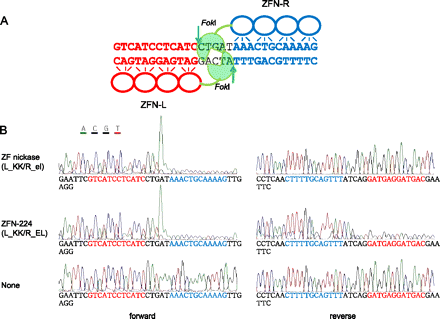
DNA cleavage by ZFNs and ZF nickases. (A) ZFN-224 and its target sequence. ZFN-224 consists of two subunits, L (left, red) and R (right, blue). The two half-site sequences are shown (red and blue) and the 5-bp spacer is shown (black). (Arrows) Cleaved phosphodiester bonds. (B) Run-off DNA sequencing analysis to detect DNA cleavage by ZFN-224 and ZF nickases. A plasmid containing the ZFN-224 target site was incubated with the nuclease pair or the nickase pair and subjected to run-off sequencing. Note that an additional adenine is added at the end by the template-independent terminal transferase activity of AmpliTaq DNA polymerase used for the sequencing reaction.











