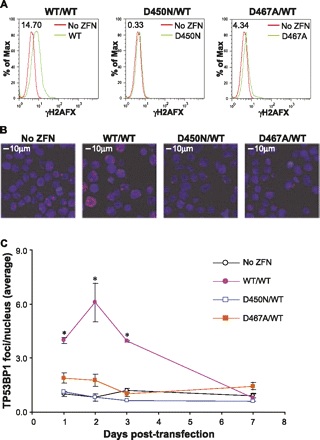
Genome-wide evaluation of DSB formation in ZFNickase-treated cells by γH2AFX and TP53BP1 staining. K562 cells were transfected with the CCR5-patch donor in the absence (No ZFN) or presence of the indicated ZFN/ZFNickase combinations. Cells were collected at 2 d (A,B) or at the indicated time (C) post-transfection and subjected to anti-γH2AFX (A) or anti-TP53BP1 (B,C) antibody staining for flow cytometry analysis (A) or immunofluorescence microscopy (B,C). Numbers of TP53BP1+ loci were counted using the SimplePCI6 software (Compix) and expressed as average TP53BP1+ foci/nucleus (C) based on counting of at least three randomly selected fields. (*) A more than twofold difference in average TP53BP1+ foci/nucleus and p < 0.05 in comparison to the non-ZFN-treated control sample (Student's t-test).











