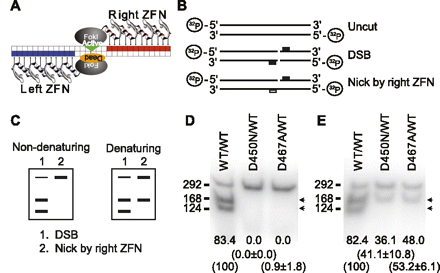
Mutation of the catalytic domain of a ZFN monomer generates a ZFN heterodimer with nicking activity. (A) Illustration of the ZFN heterodimer architecture for single-stranded break (SSB) generation. The FokI catalytic domain of ZFN (left, blue) is mutated to generate a catalytically inactivated form. This mutant productively heterodimerizes with the WT ZFN (right, red), resulting in cleavage of a single DNA strand. (B) Illustration of the DNA products following in vitro cleavage with the indicated WT and mutant ZFN combinations relative to the 5′ end-labeled DNA substrate. The position of the WT FokI ZFN binding site(s) is shown as a raised solid block and the anticipated cleavage events indicated. (C) Illustration of the expected digestion patterns following strand-specific nicking when resolved under nondenaturing and denaturing conditions. Strand-specificity is revealed since the cleavage site is off-center and the DNA substrate is labeled at the 5′ end only (see panel B above). (D) In vitro assessment of double-strand DNA cleavage with the D450N/WT and D467A/WT ZFN variants under nondenaturing conditions. (E) In vitro assessment of nicking activity with the D450N/WT and D467A/WT ZFN variants under denaturing conditions. The WT/WT combination provides a positive control for nicking of both strands. Numbers at the bottom of each lane indicate % of cleaved products (indicated by arrows). Numbers in parentheses shown are % cleavage activity (Ave ± SD) relative to the WT/WT combination (set as 100%).











