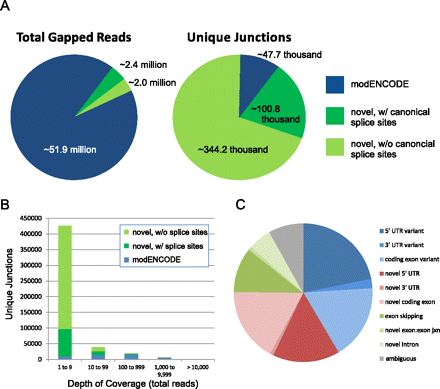
Novel splicing events in the brain transcriptome. (A) The majority of gapped reads map to previously annotated splice junctions (left); however, the vast majority of unique splice junctions detected in the brain transcriptome have not been previously annotated (right). (Blue) Splice junctions previously detected by the modENCODE Consortium (Graveley et al. 2011). (Green) Novel splice junctions with (dark green) or without (light green) canonical 5′ and 3′ acceptor/donor splice sites. (B) The depth of coverage of unique splicing events is plotted as a histogram. Not surprisingly, the more abundant a splice junction, the more likely it has been previously annotated. (C) The molecular identity of novel splicing events within ion channel genes was manually curated, with relative abundance plotted as a pie chart.











