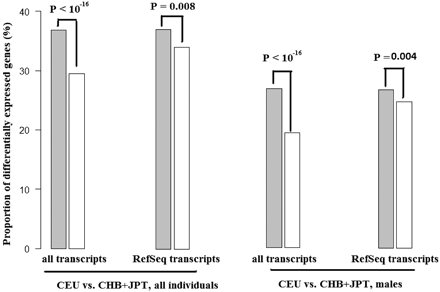
High-confidence miRNA target genes tend to display greater differential expression between CEU vs. Asian populations (FDR is cut off at 0.001). This pattern holds true either when we incorporate all of the probes or restrict our analysis to the RefSeq transcripts. Similar patterns were observed when we considered both genders, or males alone. High-confidence miRNA targets were considered as miRNA targets, while genes not targeted by coexpressed miRNAs in all of the six methods were considered non-targets (see legend to Fig. 2). (Gray) miRNA targets; (white) non-targets. When we only consider RefSeq transcripts, 3187 of 8660 (36.8%) miRNA target transcripts are differentially expressed, and 818 of 2421 (33.8%) non-target transcripts are differentially expressed (P = 0.007). For all of the expressed transcripts in males, 26.9% (2332 out of 8684) of targets are differentially expressed, and 19.4% (1080 out of 5560) of non-targets are differentially expressed (P < 10−16). For the RefSeq data in males, 25.6% (2217 out of 8660) of targets are differentially expressed, and 22.7% (550 out of 2421) of non-targets are differentially expressed (P < 10−16).











