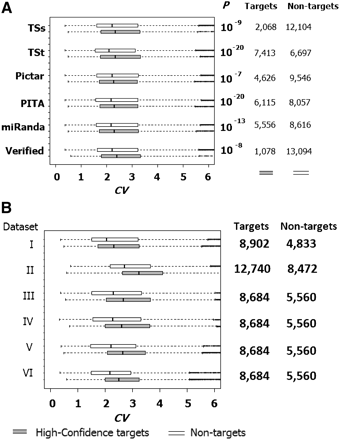
Expression variability (CV) in the lymphoblastoid cells is significantly higher for genes that are targeted by the coexpressed 287 miRNAs, regardless of target determination methods. (A) The results for data set I (Wang et al. 2009); (TSs) TargetScan with conservation (PCT > 0.8); (TSt) context score (≤0.4 and percentile >85); (Verified) experimentally verified targets taken from TarBase, miR2Disease, and Hafner et al. (2010). (B) The results after combining miRNA target determination methods together. (High-confidence miRNA targets) Only targets determined by TSs (PCT > 0.8); (TSt) (≤0.4 and percentile >85), Pictar, or experimentally verified targets of the 287 coexpressed miRNAs; (Non-targets) transcripts that are not targeted by the 287 coexpressed miRNAs with any of the miRNA target determination methods (including the four aforementioned methods, PITA, and miRanda algorithms). Boxplots of CVs for targets (gray) and non-target transcripts (white) are presented.











