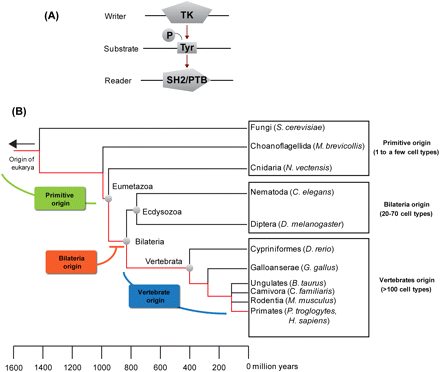
A schematic drawing of the tyrosine kinase (TK) signaling circuit and grouping of species used for the analysis of the human TK circuit evolution. (A) A simplified TK circuit that comprises a kinase that functions as the “writer,” a substrate that contains a specific tyrosine (Tyr) site for phosphorylation, and an SH2 or PTB domain-containing protein that functions as the reader of the phosphorylated Tyr (pTyr). (B) A phylogenetic tree of representative species included in the analysis of the evolution of human TK circuit evolution. They were classified into three groups. (1) The primitive group, including S. cerevisiae, M. brevicollis, and N. vectensis, represents organisms branched from the human lineage before the emergence of bilaterians. (2) Ecdyosozoans, including C. elegans and D. melanogaster, represent organisms between the branches of primitive organisms and vertebrates. (3) Vertebrates that contain the vertebrate animals we analyzed in this study, representing different branches of vertebrate evolution. The human ancestry line is shown in red. Based on this grouping, we defined origins of human protein orthologs as follows. If an ortholog is found in a primitive organism, it is assigned a primitive (or P)-origin. Similarly, if an ortholog is identified in an ecdyosozoan organism but not a primitive organism, it is considered to have originated from the bilateria (or B-origin). Orthologs found only in vertebrates are assigned a V-origin. The branch times were estimated based on data from the TimeTree server (Hedges et al. 2006).











