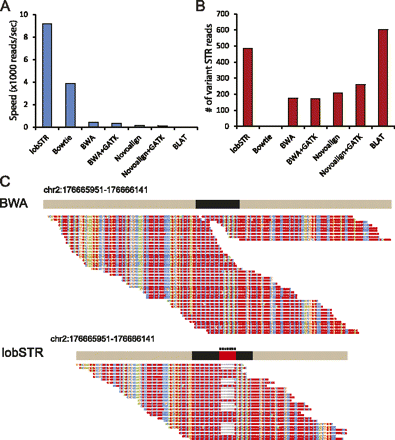
lobSTR shows an added value for STR profiling over mainstream techniques. (A) Alignment speed (reads per second) of lobSTR, mainstream aligners, and BLAT. lobSTR processes reads between 2.5 and 1000 times faster than alternative methods. (B) The sensitivity of detecting STR variations of different alignment strategies. Only BLAT detected more STR variations than lobSTR. (C) lobSTR accurately detects pathogenic trinucleotide expansions that are discarded by mainstream aligners. The figure shows simulation results of the HOXD13 heterozygous locus with a normal and a pathogenic allele that contains seven additional alanine insertions. BWA reports only the normal allele. Reads exhibiting a pathogenic STR expansion are not detected. lobSTR identifies both alleles present at the simulated locus. All positions are according to hg18.











