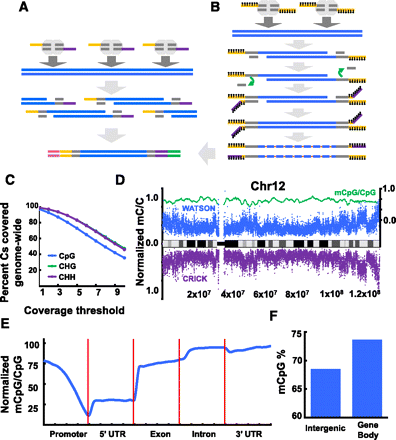
The Tn5mC-seq method and resulting methylation profiles. (A) Tagmentation-based DNA-seq library construction. Genomic DNA is attacked by transposase homodimers loaded with synthetic, discontinuous oligos (yellow, purple) that allow for fragmentation and adaptor incorporation in a single step. Subsequent PCR appends outer flowcell-compatible primers (pink, green). (B) Tn5mC-seq library construction. Loaded transposase attacks genomic DNA with a single methylated adaptor (yellow). An oligo-replacement approach anneals a second methylated adaptor (purple), which is then subject to gap-repair. Bisulfite treatment then converts unmethylated cytosine to uracil (orange) followed by PCR to append outer flowcell-compatible primers (pink, green). Methylation is represented as black lollipops. (C) Coverage of cytosine positions genome-wide. More than 96% of Cs in all three contexts are covered at least once. Slight decrease in CpG coverage is due to reduced read alignment ability at regions with a high density of methylation. (D) Normalized methylated cytosine over total cytosine positions in 10-kb windows across chromosome 12 (blue and purple, left axis), and normalized methylated CpG over total CpG in 100-kb windows across chromosome 12 (green, right axis). (E) Normalized methylated CpG over total CpG residues at annotated genic loci. Promoter is defined as 2-kb region upstream of TSS. (F) Elevated CpG methylation levels in gene body (intron, exon) compared to intergenic regions.











