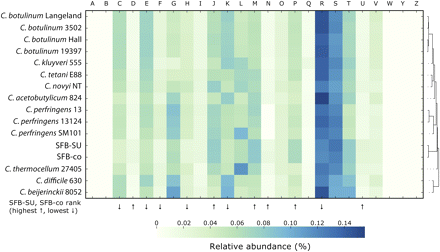
Functional categories of cluster orthologous groups (COGs). Analysis of the predicted proteome of SFB-co and SFB-mouse-SU in comparison to other clostridia. Relative abundance levels of COG functional categories were clustered using a Euclidean distance metric and displayed as a heatmap, with low abundance (light green) and high abundance (dark blue). (A) RNA processing and modification. (B) Chromatin structure and dynamics. (C) Energy production and conversion. (D) Cell cycle control, cell division, chromosome partitioning. (E) Amino acid transport and metabolism. (F) Nucleotide transport and metabolism. (G) Carbohydrate transport and metabolism. (H) Coenzyme transport and metabolism. (I) Lipid transport and metabolism. (J) Translation, ribosomal structure and biogenesis. (K) Transcription. (L) Replication, recombination and repair. (M) Cell wall/membrane/envelope biogenesis. (N) Cell motility. (O) Post-translational modification, protein turnover, chaperones. (P) Inorganic ion transport and metabolism. (Q) Secondary metabolites biosynthesis, transport, and catabolism. (R) General function prediction only. (S) Function unknown. (T) Signal transduction mechanisms. (U) Intracellular trafficking, secretion, vesicular transport. (V) Defense mechanisms. (W) Extracellular structures. (Y) Nuclear structure. (Z) Cytoskeleton.











