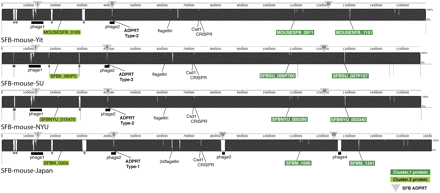
Genome sequence comparison of whole and nearly complete SFB genomes, and SFB-specific proteins. The genome sequences of SFB-mouse-SU, SFB-mouse-Yit (AP012209) (Prakash et al. 2011), SFB-mouse-NYU (AGAG01000000) (Sczesnak et al. 2011), and SFB-mouse-Japan (AP012202) (Kuwahara et al. 2011) were aligned using Mauve. (Black-filled areas) High sequence identity; (white-filled areas) differences among some or all genomes. SFB-specific proteins that exhibit extensive polymorphisms among genomes are labeled: Cluster.1 proteins (dark green box); Cluster.3 proteins (light green box). (Triangles) The genome locations of the four types of ADP-ribosyltransferases. (*) Ribosomal RNA operons that are missing from some genomes. Thin vertical black lines for SFB-mouse-SU, and SFB-mouse-NYU indicate the borders of contigs. Two similar flagellins (FliC3, and FliC4) in SFB-mouse-Japan are encoded at a position where only one copy is observed in the other genomes; sequence fragments from single cell SFB suggest the presence of two flagellins as well (this study). See Supplemental Figure S9 and Supplemental Table S3 for further sequence comparisons of SFB-specific proteins among genomes.











