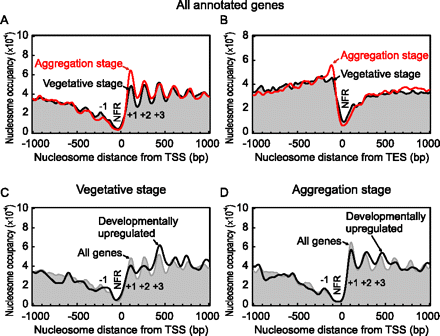
Nucleosome organization around the 5′ and 3′ end of the Dictyostelium genes. (A,B) Composite distribution of in vivo nucleosome positions relative to the TSS and TES. The midpoints of all nucleosomal sequence reads were distributed around the TSS (or TES) of 5468 (or 5400) protein-coding genes, plotted as described in Figure 1B and smoothed with a bandwidth of 15 bp. The free-living (vegetative) and multicellular (aggregation) stages are indicated by black and red traces, respectively. (C,D) Nucleosome organization around the TSS of 325 developmentally up-regulated genes (black trace) in the vegetative and aggregation stages, compared to all nucleosomes (gray fill). Traces were smoothed with a 30-bp bandwidth.











