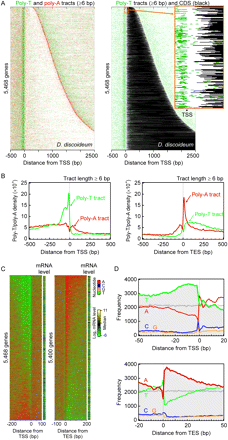
Dictyostelium gene organization with poly-T/poly-A enrichment. (A) Distribution of poly-T/poly-A tracts (≥6 bp) in 5468 genes with the annotated TSS. Each track represents the DNA sequence of a gene, fetched from −500 to 2500 relative to the TSS. Genes were aligned by their TSS, and their DNA sequence in the sense strand was oriented in the 5′ to 3′ direction. If any poly-T/poly-A tract of length six or more was found in the DNA sequences, the poly-T sequence was colored in green and the poly-A sequence in red, and all the others were drawn as white in the sequence. The genes were sorted in ascending order of length. The right panel shows poly-T tracts as green and coding sequences as black. A zoom-in screenshot demonstrates the positional details in poly-T enrichment at TSS and translation start sites. (B) Composite distribution of the location of poly-T/poly-A tracts around the 5′ and 3′ end of Dictyostelium genes. The density distribution of poly-T/poly-A tracts (≥6 bp) was displayed as a function of the distance (bp) on the sense strand between the midpoint of each nonoverlapping tract and the given TSS (or TES). The density of the occurrence of poly-T (green trace) and poly-A tracts (red trace) is shown in the y-axis, which was estimated by Gaussian kernel and a smoothing bandwidth of 5. The density curve was calculated within the range from −1500 to 1500 relative to the TSS (or TES) and only the −500 to 500 region shown in the figure. (C) Transcription start (TSS) and end (TES) site are linked to high T and A enrichment, respectively, in the adjacent intergenic sequence. Each track represents the sense strand of a gene, fetched from 200 bp intergenic to 100 genic relative to the TSS (or TES) of 5468 (or 5400) protein-coding genes. Genes are aligned by the TSS (left) or TES (right), in the 5′ to 3′ direction from left to right. Transcript abundance is shown for each gene in an adjacent column. Genes were ordered by descending T (left) or A (right) density of their extracted sequence (301 bp). Color codes for the four nucleotides are indicated. This trend was not observed with randomization simulations (not shown). (D) Frequency distribution of A, C, G, and T relative to the TSS and TES. Shown is a summation of columns from panel C, over the indicated distance, color-coded as in panel C. The same simulation was applied to a set of TSSs or TESs randomly positioned across the Dictyostelium genome (gray traces, shown for A and T).











