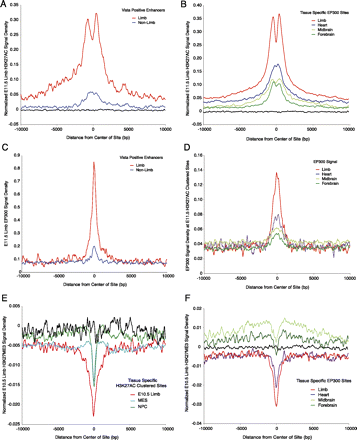
Active and inactive enhancers are marked by both EP300 and H3K27ac. (A) E11.5 limb H3K27ac signal density normalized for input was determined in a 20-kb window surrounding the center of Vista limb enhancers or positive enhancers with no limb annotation. The strongest signal and nucleosome displacement are observed at known limb positive enhancers. Active enhancers with no observed limb activity are marked with low levels of H3K27ac. Black lines represent signal at random intergenic and intronic regions. (B) E11.5 limb H3K27ac signal density normalized for input at tissue-specific intergenic and intronic EP300 sites. Signal is strongest at limb-specific EP300 sites and shows a strong signal of H3K27ac modified nucleosome displacement. Other tissue-specific EP300 sites are enriched for H3K27ac compared with random regions (black), but at lower levels. (C) Limb EP300 signal density at Vista limb enhancers or positive enhancers with no limb annotation. Signal was normalized versus random regions since no input was performed for these experiments. Limb EP300 signal is strongest at known limb enhancers, but is also present at low levels at enhancers with no limb activity. (D) EP300 signal density from four mouse tissues at E11.5 H3K27ac clustered sites. Signal was normalized versus random regions as in C. EP300 signal from limb is strongest at limb-specific H3K27ac regions, but EP300 signal from other tissues is also present at limb-specific H3K27ac regions. (E) E10.5 limb H3K27me3 signal density normalized for input at tissue-specific H3K27ac intronic and intergenic regions identified in this study. All classes show depletion of H3K27me3 versus random genomic regions (black), but limb-specific regions are most depleted. (F) E10.5 limb H3K27me3 signal density normalized for input at tissue-specific intergenic and intronic EP300 sites. All classes show depletion of H3K27me3 relative to their surrounding genomic locations. Limb and heart-specific sites exhibit depletion versus random genomic regions (black).











