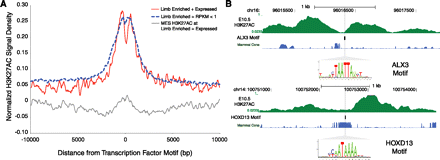
Strongly marked limb H3K27ac regions are enriched for limb transcription-factor binding sites. (A) E10.5 limb H3K27ac signal density plots normalized for input in a 20-kb window centered on limb-enriched transcription-factor (TF) motifs (Methods) for factors expressed highly in limb versus those that are expressed at low levels (RPKM <1). Those TF motifs that are enriched in limb and whose corresponding factors are highly expressed show signatures of H3K27ac modified nucleosome displacement (red). Motifs enriched in limb, but whose factors are expressed at low levels do not exhibit nucleosome displacement (blue). MES H3K27ac signal at limb-enriched motifs for TFs highly expressed in limb shows low signal and no nucleosome displacement (gray). (B) Displacement of H3K27ac signal in H3K27ac-enriched regions relative to the presence of predicted binding sites for Alx3 (top) and Hoxd13 (bottom).











