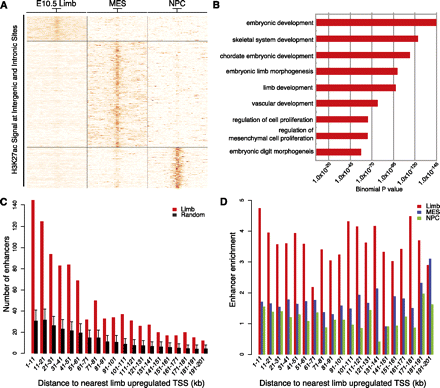
Tissue-specific H3K27ac intergenic and intronic regions are associated with genes up-regulated in E10.5 limb. Intergenic and intronic H3K27ac regions in limb, MES, and NPC were identified and merged as described in the main text. (A) K-means clustering of H3K27ac signals (in RPKM) across 39,750 merged regions, using an 8-kb window centered on the midpoint of each region, revealed three tissue-specific classes of H3K27ac-enriched regions. (B) Selected GO Biological Process category enrichments identified by GREAT analysis of “limb-specific” H3K27ac regions. (C) Counts of limb-specific H3K27ac regions (red) relative to random sites (black) near genes up-regulated in limb. Each region was assigned to the nearest transcription start site up to 200 kb away, and enrichment of limb-specific regions near limb up-regulated genes was calculated in 10-kb bins. Error bars represent the 95% quartile of values from 1000 randomized sets of intergenic and intronic sequences. (D) Enrichment of assigning a limb (red), MES (blue), or NPC-specific region (green) to a limb up-regulated gene versus random regions used in C up to 200 kb away.











